Inferring the Causal Role of Aging-Related Factors in MS Progression
A bidirectional Mendelian Randomization analyses of mitochondrial DNA copy number and leukocyte telomere length in multiple sclerosis progression.
Status: Published · Category: Research / Mendelian Randomization · Includes: Bidirectional MR (mtDNA-CN ↔ MS progression), Bidirectional MR (LTL ↔ MS progression), reproducible HTML reports, published papers.
1. Statement of the Problem and Rationale
Multiple Sclerosis (MS) is a chronic inflammatory disease of the central nervous system characterized by progressive neuronal loss that primarily affects young adults (1). The pathogenesis involves the dysregulation of both adaptive and innate immune cells targeting CNS autoantigens, which drives chronic neuroinflammation and subsequent neuronal damage (2). The clinical course is variable; most patients are diagnosed with relapsing-remitting MS (RRMS), defined by episodes of acute neurological dysfunction followed by recovery. Over time, however, a majority transition to secondary progressive MS (SPMS), a stage of continuous neurodegeneration without distinct relapses. A smaller subset (~10-15%) experience primary progressive MS (PPMS), with a steady accumulation of disability from onset (3). This transition from inflammation-driven relapses to relentless neurodegeneration represents the core clinical challenge in MS and an area of urgent unmet need. Biological aging and associated cellular dysfunction are recognized as critical drivers of MS progression. Two key biomarkers of these processes are mitochondrial DNA copy number (mtDNA-CN) and leukocyte telomere length (LTL) (4-6).
mtDNA-CN is a critical indicator of cellular bioenergetic health. Mitochondria play central roles in cellular proliferation, maintaining mitochondrial membrane potential, regulating apoptosis, managing oxidative stress, storing energy, and producing adenosine triphosphate (ATP). Fluctuations in mtDNA-CN signal potential bioenergetic deficits and impaired respiratory capacity in neurons, which may directly contribute to the neuronal loss seen in MS (7-9).
(LTL) is a well-established biomarker of biological aging. Telomeres are nucleoprotein structures that protect the ends of chromosomes, regulating cellular senescence and maintaining genomic stability (6). Observational studies have linked shorter LTL to progressive MS forms (10), higher disability scores, and reduced brain volume (11), suggesting accelerated biological aging contributes to disease severity.
Despite these associations, prior observational research has yielded inconsistent findings and is methodologically incapable of establishing causality. These studies are highly susceptible to confounding variables and reverse causation (12), making it impossible to determine if alterations in mtDNA-CN and LTL are a cause of MS progression or merely a consequence of the disease process. This ambiguity has stalled progress in understanding the fundamental mechanisms of neurodegeneration in MS, creating a critical barrier to developing effective, targeted therapies.
To overcome these limitations, this study will employ Mendelian Randomization (MR), a robust genetic epidemiology method that uses randomly inherited genetic variants as instrumental variables. Analogous to a randomized controlled trial, MR mitigates confounding and allows for the inference of causal directionality (13). This project outlines a methodologically rigorous investigation designed to leverage the power of MR to dissect the causal roles of mtDNA-CN and LTL in the progression of multiple sclerosis.
2. Research Objectives
The overarching goal of this research is to dissect the causal relationships between key biomarkers of cellular function and biological aging, namely, mtDNA-CN and LTL, and the progression of MS. By employing a genetically-informed MR framework, this study will move beyond mere association to establish the directionality and causality of these critical biological pathways, addressing a fundamental gap in our understanding of MS pathophysiology. The primary and secondary objectives of this study are:
- Objective 1: To Investigate the causal effect of mtDNA-CN on MS progression. To determine if genetically predicted variation in mtDNA-CN has a causal impact on the age-related severity of MS.
- Objective 2: To investigate the causal effect of leukocyte telomere length on MS progression. To determine if genetically predicted LTL has a causal impact on the age-related severity of MS.
- Objective 3: To assess for reverse causality. Utilizing a bidirectional MR framework, this aim will test the alternative hypothesis: whether genetically predicted MS progression causally influences levels of mtDNA-CN and LTL, thereby clarifying the direction of the biological relationship.
3. Methodological Framework
This research will employ a bidirectional, two-sample MR design. This framework is strategically chosen for its statistical power and its ability to infer both causality and directionality by leveraging summary-level data from large-scale, publicly available genome-wide association studies (GWAS). This approach enables a rigorous investigation while obviating the need for new patient recruitment and data collection.
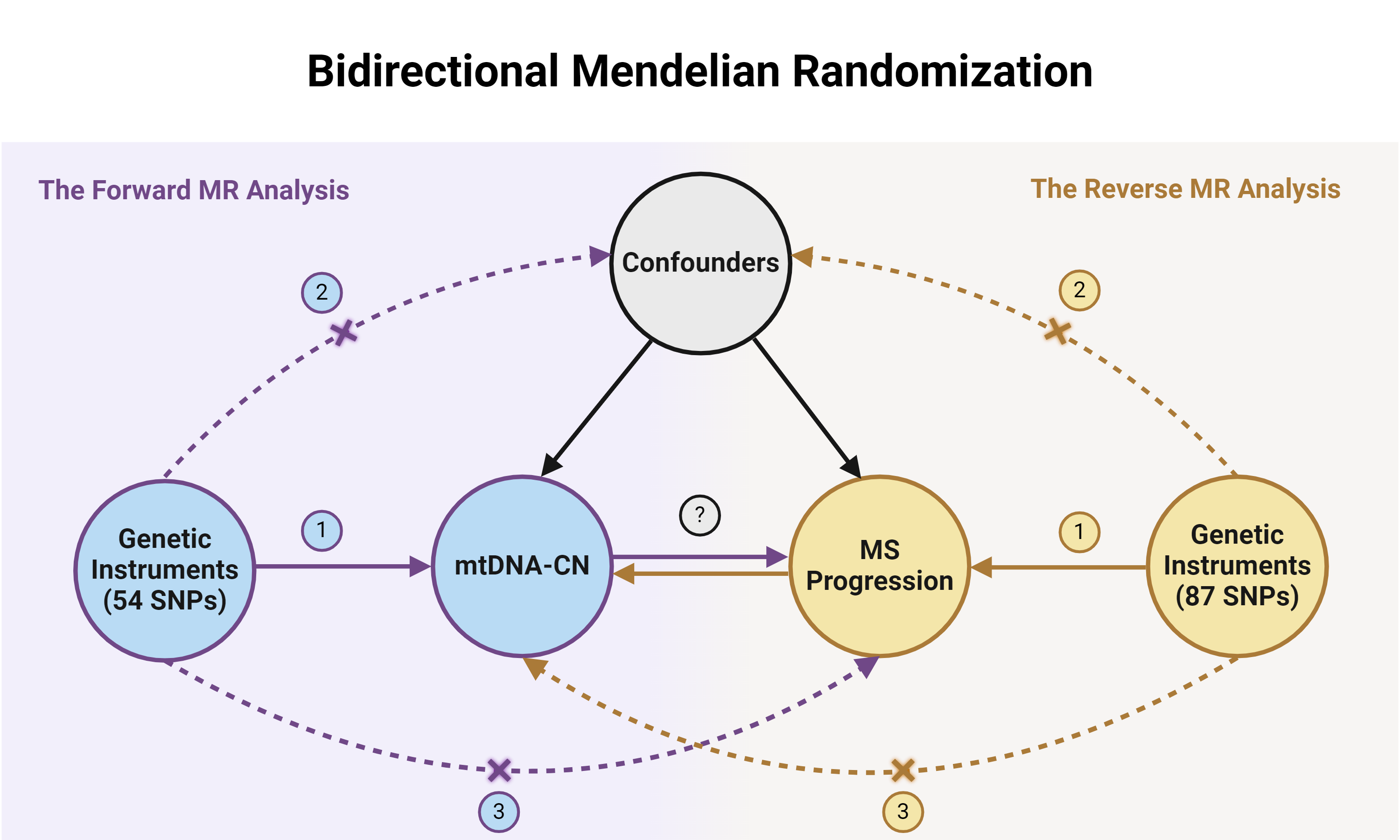
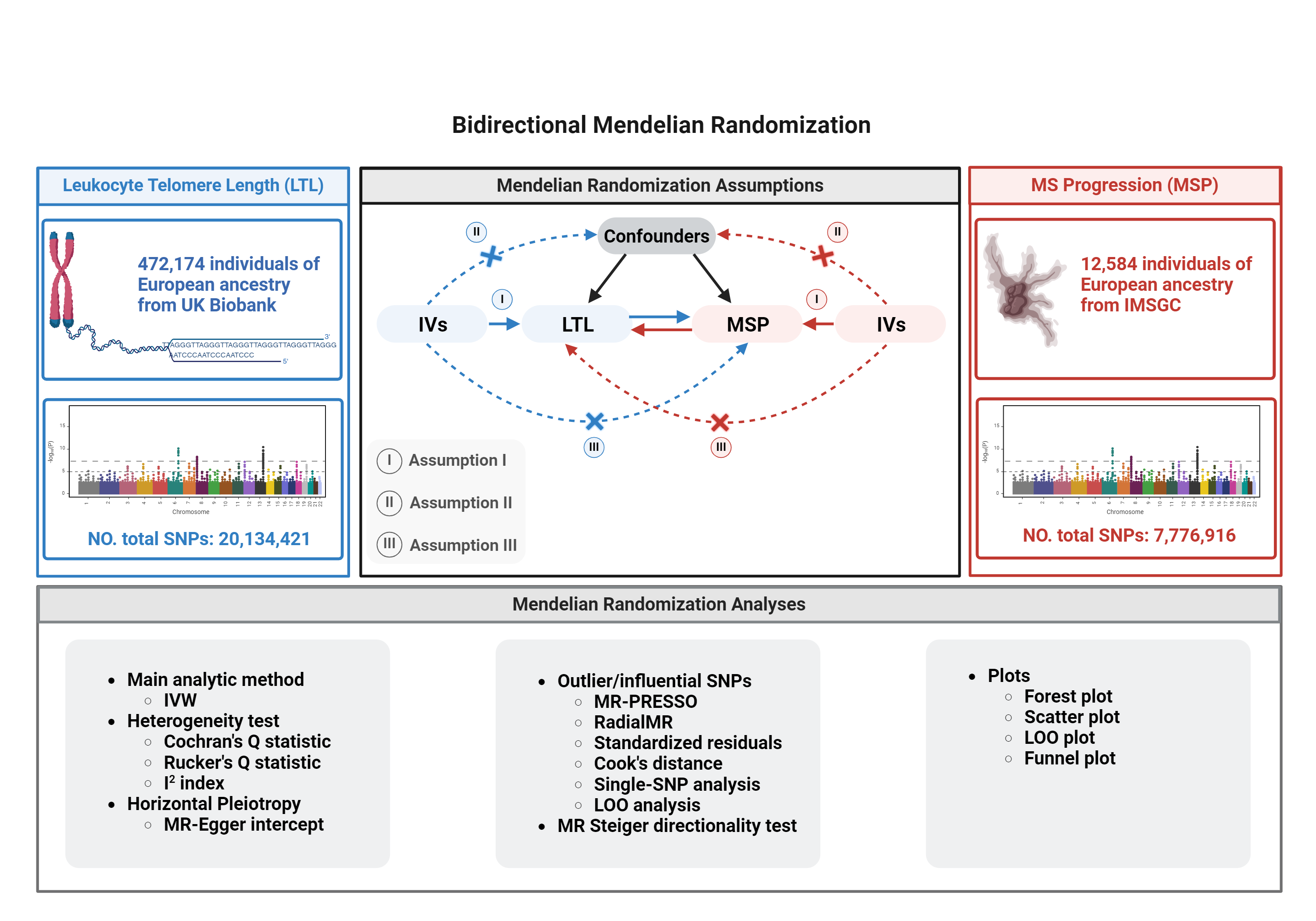
3.1 Data Sources
This study will utilize summary-level genetic data from large-scale GWAS consortia. The details of these datasets, which include hundreds of thousands of individuals of European ancestry, are summarized below.
| Trait | Resource | Sample Size (European Ancestry) | Key Covariates |
|---|---|---|---|
| Mitochondrial DNA Copy Number (mtDNA-CN) | UK Biobank | 383,476 | Blood cell counts, 20 genetic principal components, age, sex, and chip type |
| Leukocyte Telomere Length (LTL) | UK Biobank | 472,174 | Adjusted for methodological factors, and associations with established LTL-related phenotypes, including sex, ethnicity, and age, were confirmed |
| Age-Related MS Severity Score (ARMSS) | International Multiple Sclerosis Genetics Consortium (IMSGC) | 12,584 | Sex, age at onset, date of birth, center, genotyping platform, and the first ten principal components |
3.2 Genetic Instrument Selection and Validation
A rigorous process will be used to select and validate the single-nucleotide polymorphisms (SNPs) that will serve as instrumental variables (IVs) for the MR analysis.
- SNP Selection: SNPs associated with the exposures (mtDNA-CN and LTL) will be selected based on a genome-wide significance threshold of P < 5 × 10–8, P < 5 × 10–6, or P < 5 × 10–5.
- Instrument Validity: To ensure that the selected IVs meet the core assumptions of MR, the following validation steps will be performed:
- Linkage Disequilibrium: To ensure the independence of genetic instruments, SNPs will be clumped using a strict threshold of r2 < 0.001 within a 10,000 kb genomic window.
- Instrument Strength: Weak instrument bias will be minimized by calculating the F-statistic for each SNP and excluding any with a value less than 10 (F < 10).
- Data Harmonization: The effect alleles for each SNP will be harmonized across exposure and outcome datasets. Palindromic SNPs with ambiguous allele frequencies will be excluded to prevent analytical errors.
3.3 Statistical Analysis Plan
The causal effect of each exposure on the outcome will be estimated using a comprehensive and multi-faceted statistical framework to ensure maximum robustness.
- The Inverse Variance Weighted (IVW) method will serve as the primary analysis technique for estimating the overall causal effect.
- To rigorously test MR assumptions and ensure the stability of our findings, an exhaustive suite of sensitivity and secondary analyses will be deployed. These include multiple methods robust to violations of standard MR assumptions:
- Pleiotropy-Robust Methods: A full range of techniques will be applied, including MR-Egger, Penalized MR-Egger, Robust MR-Egger, Simple Median, Weighted Median, Penalized Weighted Median, Simple Mode, Weighted Mode, Robust Adjusted Profile Score (RAPS), MR-Constrained Maximum Likelihood (MR-cML), Debiased IVW (dIVW), Mode-Based Estimation (MBE), and MR-Lasso.
- Heterogeneity and Pleiotropy Assessment: Heterogeneity will be evaluated using Cochran’s Q, Rucker’s Q statistic, and the I² index. Directional horizontal pleiotropy will be formally assessed using the MR-Egger intercept test.
- Outlier Detection and Correction: Potential pleiotropic outliers will be identified and corrected using multiple methods, including the Mendelian Randomization Pleiotropy RESidual Sum and Outlier (MR-PRESSO) test, RadialMR, Cook’s distance, and standardized residuals.
- Influence Analysis: A Leave-One-Out (LOO) analysis will be performed to systematically evaluate whether the overall causal estimate is being disproportionately influenced by any single genetic variant.
- The MR Steiger directionality test will be applied to statistically confirm the assumed causal direction from exposure to outcome. All statistical analyses will be performed using R software (V.4.3.2) with the established packages “TwoSampleMR,” “MR-PRESSO,” “MendelianRandomization,” “MRPracticals,” and “mr.raps”. An association with a P < 0.05 will be considered statistically significant.
4. Expected Outcomes and Scientific Significance
By moving beyond simple association to establish causality, this study is poised to deliver critical new insights into the biological mechanisms that drive the progression of MS. The findings from this methodologically rigorous investigation have the potential for significant scientific and clinical impact. The expected outcomes include:
- Causal Evidence: This study will provide genetically-validated evidence to determine whether mitochondrial dysfunction (via mtDNA-CN) and accelerated biological aging (via LTL) are causal drivers of neurodegeneration in MS. The bidirectional design will definitively clarify whether these biomarker changes are a cause of disease progression or merely a consequence of it.
- Identification of Novel Therapeutic Targets: If a causal link is established, these biomarkers would represent novel and highly promising targets for therapeutic intervention. Developing therapies that modulate mitochondrial function or slow cellular aging could offer a new frontier for treatments aimed at halting the accumulation of disability, a major area of unmet clinical need.
- Improved Understanding of Pathophysiology: The results will significantly advance our fundamental understanding of MS pathology. Whereas previous genetic studies have primarily focused on variants influencing MS susceptibility, this research will delineate the distinct genetic architecture and biological pathways that influence disease progression, clarifying why disability accumulates more rapidly in some individuals than in others.
Reproducible Analysis Reports


Interactive HTML reports:
Related Publications
-
Sabaie H, Taghavi Rad A, Shabestari M, Habibi D, Saadattalab T, Seddiq S, Saeidian AH, Zahedi AS, Sanoie M, Vahidnezhad H, Zarkesh M, Foroutani L, Hakonarson H, Azizi F, Hedayati M, Daneshpour MS, Akbarzadeh M. Mitochondrial DNA Copy Number as a Hidden Player in the Progression of Multiple Sclerosis: A Bidirectional Two-Sample Mendelian Randomization Study. Molecular Neurobiology. 2025;62(9):11643-53.
https://doi.org/10.1016/j.msard.2025.106277 -
Sabaie H, Taghavi Rad A, Shabestari M, Seddiq S, Saadattalab T, Habibi D, Saeidian AH, Abbasi M, Mirtavoos-Mahyari H. Deciphering the bidirectional impact of leukocyte telomere length on multiple sclerosis progression: A Mendelian randomization study. Multiple Sclerosis and Related Disorders. 2025;94:106277.
https://doi.org/10.1016/j.msard.2025.106277
References
- Friese MA, Schattling B, Fugger L. Mechanisms of neurodegeneration and axonal dysfunction in multiple sclerosis. Nat Rev Neurol. 2014;10(4):225-38.
- Ellwardt E, Zipp F. Molecular mechanisms linking neuroinflammation and neurodegeneration in MS. Exp Neurol. 2014;262 Pt A:8-17.
- Doshi A, Chataway J. Multiple sclerosis, a treatable disease. Clin Med (Lond). 2016;16(Suppl 6):s53-s9.
- Bektas A, Schurman SH, Sen R, Ferrucci L. Aging, inflammation and the environment. Exp Gerontol. 2018;105:10-8.
- Graves JS, Krysko KM, Hua LH, Absinta M, Franklin RJM, Segal BM. Ageing and multiple sclerosis. Lancet Neurol. 2023;22(1):66-77.
- Blackburn EH, Epel ES, Lin J. Human telomere biology: A contributory and interactive factor in aging, disease risks, and protection. Science. 2015;350(6265):1193-8.
- Gustafsson CM, Falkenberg M, Larsson NG. Maintenance and Expression of Mammalian Mitochondrial DNA. Annu Rev Biochem. 2016;85:133-60.
- Guha M, Avadhani NG. Mitochondrial retrograde signaling at the crossroads of tumor bioenergetics, genetics and epigenetics. Mitochondrion. 2013;13(6):577-91.
- Cerantonio A, Citrigno L, Greco BM, De Benedittis S, Passarino G, Maletta R, Qualtieri A, Montesanto A, Spadafora P, Cavalcanti F. The Role of Mitochondrial Copy Number in Neurodegenerative Diseases: Present Insights and Future Directions. Int J Mol Sci. 2024;25(11).
- Guan JZ, Guan WP, Maeda T, Guoqing X, GuangZhi W, Makino N. Patients with multiple sclerosis show increased oxidative stress markers and somatic telomere length shortening. Mol Cell Biochem. 2015;400(1-2):183-7.
- Krysko KM, Henry RG, Cree BAC, Lin J, Caillier S, Santaniello A, Zhao C, Gomez R, Bevan C, Smith DL, Stern W, Kirkish G, Hauser SL, Oksenberg JR, Graves JS. Telomere Length Is Associated with Disability Progression in Multiple Sclerosis. Ann Neurol. 2019;86(5):671-82.
- Grimes DA, Schulz KF. Bias and causal associations in observational research. The Lancet. 2002;359(9302):248-52.
- Sekula P, Del Greco MF, Pattaro C, Köttgen A. Mendelian Randomization as an Approach to Assess Causality Using Observational Data. J Am Soc Nephrol. 2016;27(11):3253-65.